Few Exemplar-Based General Medical Image Segmentation via Domain-Aware Selective Adaptation
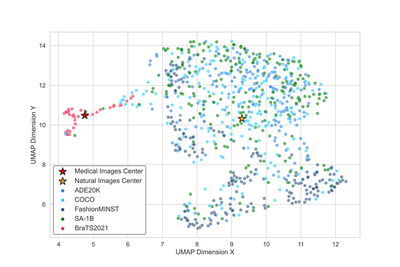
The dimensionality reduction visualisation map shows the relative relationship and distribution of different data sets in a two-dimensional feature space. It can be seen the characteristics of the medical data set (red points) compared to the general natural data set (blue/black points). A clear domain gap can be observed from this visualisation.
Chen Xu†, Qiming Huang†, Yuqi Hou, Jiangxing Wu, Fan Zhang, Hyung Jin Chang, Jianbo Jiao
The MIx Group, University of Birmingham, Fudan University
 FEMed Architecture
FEMed ArchitectureThe architecture of our Image Encoder enhanced with two specialized Adapters: (a) the Multi-Scale Features Adapter that captures features at various granularities through pyramid pooling, and (b) the High-Frequency Adapter that emphasizes salient textural details from frequency domain analysis. (c) These Adapters feed into the Selection Module, which uses a trainable binary decision layer to selectively integrate the most informative feature set at each transformer stage, effectively tailoring the feature landscape for optimal tumour delineation in CT/MRI scans.
- Yang, Lingfeng, Wang, Yueze, Li, Xiang, Wang, Xinlong, Yang, Jian. "Fine-grained visual prompting." Advances in Neural Information Processing Systems, vol. 36, 2024.
 Prompt Strategy
Prompt StrategyFour settings of using bbox prompts during training and testing stages. The coarse bounding box prompt is designed to be almost the same size as the input image data, with different ratios indicating the proportion of pixels by which the rectangle is shrunk inward relative to the entire image. A pseudo-code for coarse bbox prompt generation is shown in Algorithm 1 in the paper.
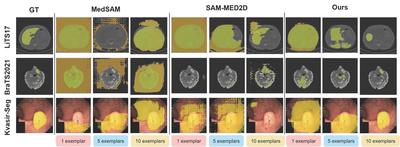
Comparison from MedSAM[2], SAM-MED2D[3], and our FEMed method (5-shot, 10-shot). The first column is the input image, the second column is the image with coloured ground truth masks, and the third and fourth columns are the image with coloured predicted masks by MedSAM and SAM-MED2D. The right two columns are the image with coloured predicted masks by our FEMed method.
- Ma, et al. "Segment Anything in Medical Images with MedSAM." 2024.
- Cheng, et al. "SAM-MED2D: Medical Image Segmentation with Segment Anything Model." 2023.